FASTQ tools
Application to show information about and scramble FASTQ files to provide non-sensitive data for development purposes
Usage
This application provides the following subcommands
Usage: fastq-tools [OPTIONS] <COMMAND>
Commands:
info Show information about input
grz-metadata Show GRZ metadata
scramble Scramble input data
help Print this message or the help of the given subcommand(s)
Options:
-i, --input <INPUT_FILE> Input file
-d, --decompress Decompress input as gzip compressed data
-h, --help Print help
-V, --version Print version
Info
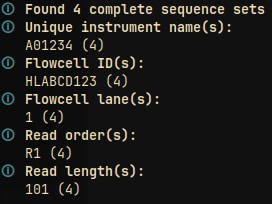
To show information about compressed FASTQ files use:
cat file_fastq.gz | gzip -d | fastq-tools info
To use build-in decompression of input data, use the --decompress/-d option:
cat file_fastq.gz | fastq-tools --decompress info
Using optional input file argument:
fastq-tools --decompress --input file_fastq.gz info
This will result in output like
GRZ Metadata
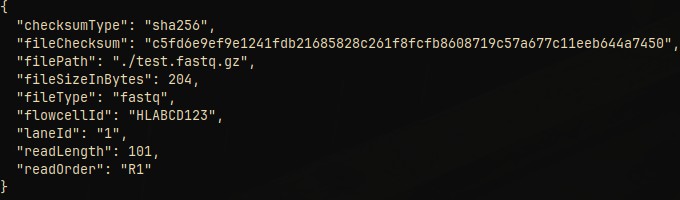
To generate GRZ metadata for a file use:
fastq-tools --decompress --input file_fastq.gz grz-metadata
The use of the --input argument is required for this sub command.
If the file is an uncompressed FASTQ file, you can omit the --decompress option.
Supported file types are:
- fastq (full support)
- bam, bed, vcf (limited support)
Scramble
To scramble compressed FASTQ files use:
cat file_fastq.gz | gzip -d | fastq-tools scramble | gzip > scrambled_fastq.gz
This will scramble headers and sequences and write the output into scrambled_fastq.gz.
To use build-in decompression of input data, use the --decompress/-d option:
cat file_fastq.gz | fastq-tools -d scramble | gzip > scrambled_fastq.gz
Using optional input file argument:
fastq-tools -d -i file_fastq.gz scramble | gzip > scrambled_fastq.gz